By Mariana Bego
New variants of the virus that causes COVID-19 are introduced almost daily. Consensus on a naming system has been difficult, so scientists are developing their own creative names for the variants to help distinguish them from one another. Variant names started to divert as fast as their diversity started to be noticed late last year creating further complications. Furthermore, terms such as ‘variant’, ‘lineage’ and ‘strain’ are often used interchangeably and they add to the overall confusion.
While one naming system indicated the evolutionary relationships between SARS-CoV-2 lineages, and called the variant originally described in the United Kingdom as B.1.1.7, others named it 20I/501Y.V1 to highlight key mutations. This variant would be known as VOC 202012/01 following WHO recommendations. Finally, seeing that these alpha-numeric names were confusing, an inspired research group appointed it “Nelly”!?
Let’s dissect the origins of the possible names for this variant as an example:
Location and dates names (The UK variant and VOC 202012/01)
This virus was originally known as the UK variant of SARS-CoV-2. Seeing as naming variants systems following the geographic area where they were first reported can generate dangerous stigmas, this name was quickly discouraged by the World Health Organization (WHO). The WHO outlined a new naming system in January of 2021, which coined the concepts of Variant Under Investigation (VUI) and Variant of Concern (VOC), followed by a reference to the month and year of discovery. Under this nomenclature this variant became VOC 202012/01.
Read more about this topic in a recent editorial in Nature.
Mutation-specific naming (N501Y and 20I/501Y.V1)
Some proposed names highlight important mutations of the variants of concern. An early name for this variant was simply N501Y reflecting how the amino acid in position 501st changed from an asparagine to a tyrosine (which are abbreviated as N and Y, respectively, following biochemical naming standards). Often variants carry more than one potentially interesting mutation, however. Scientists from Nextstrain built on this and proposed to name variants of interest after the constellations of mutations they carry. Nextstrain is an open-source project to share pathogen genome data, alongside powerful analytic and visualization tools. They were responsible for coming up with the name 20I/501Y.V1 for this particular variant. Distinct variants carrying similar sets of important mutations would earn similar names. Many scientists want to avoid names that flag individual mutations. While the name indeed allows researchers to associate the variants with the mutations that may drive their behaviours, it ignores other important changes present in the variant.
For more information visit the Nextstrain web site.
The Pango system (B.1.1.7): Rambaut and colleagues initially proposed to name variants using a dynamic nomenclature that generates technical designations such as B.1.1.7. For this designation to be valid and broadly accepted it needs to: (a) capture local and global genetic diversity; (b) track emerging lineages; (c) be robust and flexible to accommodate new diversity as it is generated; and (d) be dynamic through time. They propose to use the term ‘lineages’ (rather than ‘clades’, or ‘genotypes’) for SARS-CoV-2 because it captures the fact that they are dynamic in nature.
A phylogenetic tree is a diagram that represents evolutionary relationships among organisms, in this case the circulating SARS-CoV-2 viruses. They propose to map each sequenced virus in a phylogenetic tree and each major lineage label (or tree branch) to begin with a letter (See Figure 1). With the two main branches being lineage A viruses, represented by Wuhan/WH04/2020, and lineage B, represented by Wuhan-Hu-1 from where most the recorded human cases seem to derive to date. Rambaut and colleagues named their nomenclature system as ‘Pango’ (from the first-person Latin verb meaning ‘I set’, ‘I fix’ or ‘I record’). Recent Addendum to their original manuscript published at the end of January of 2021 provides links to several online tools for classifying SARS-CoV-2 genome sequences.
You can read their article here.
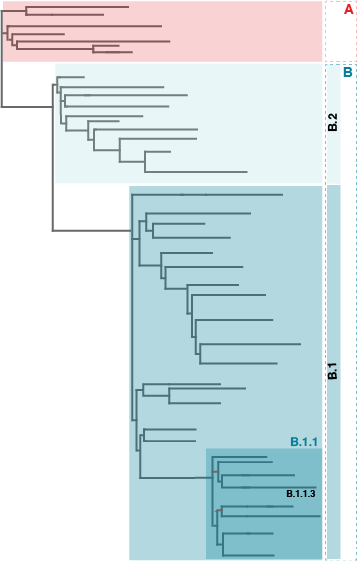
Figure 1: A phylogenetic tree used to derive names used in the Pango linages nomenclature.
You can read their article here.
Hurricane-inspired naming system (Nelly): Áine O’Toole, an evolutionary biologist at the University of Edinburgh, introduced new names in a recent interview with The Guardian, as the names mentioned above were “quite a mouthful”. In this scheme, N501Y became “Nelly”, D614G became “Doug” and E484K became “Eeek”. “It was originally going to be called Erik, but we work with someone called Eric quite a lot so we didn’t want to necessarily overlap the name of that mutation with someone we knew,” she said. To avoid upsetting friends and family, the authors of the pre-print highlighted in the blog post below, selected animal names for their provisional naming system. The names of the key variant described spiraled from the phylogenetically-inspired Pango lineages nomenclature to birds. For example, B.1.2 is now “Robin”, B.1.234 is “Mockingbird”, and B.1.1.220 is “Bluebird”. They have Pelican, Yellowhammer and Quail variants too. According to the authors of this manuscript, nicknames may work well in the short term, but are hardly a long-term solution. “And if you’re gaining all these extra birds, you’re soon going to have a zoo!” the lead author of that manuscript, Emma Hodcroft, who is also a member of Nextstrain remarked in The Guardian.
You can find the link to the article published last week in The Guardian.

